The brain consists of vast networks of nerve cells. Understanding how cells interact in such networks is crucial for developing and improving medical tools for diagnosis and treatment of brain impairments. For example, scientists are developing electrical prostheses that aim to restore functionality in damaged neural tissue, but making these prostheses more accurate requires a deeper understanding of the impaired system. In order to understand such processes in single neurons as well as interactions in networks of neurons, computational models can be used to understand the plethora of mechanisms that generate neural activity. However, identifying suitable models, i.e. models that match the measured data, has so far often been very difficult, especially for complex, mechanistic models with many parameters. In two recent papers, we developed a new method to identify suitable models and applied it to a computational model of retinal prostheses – implants which aim to restore vision in the blind.
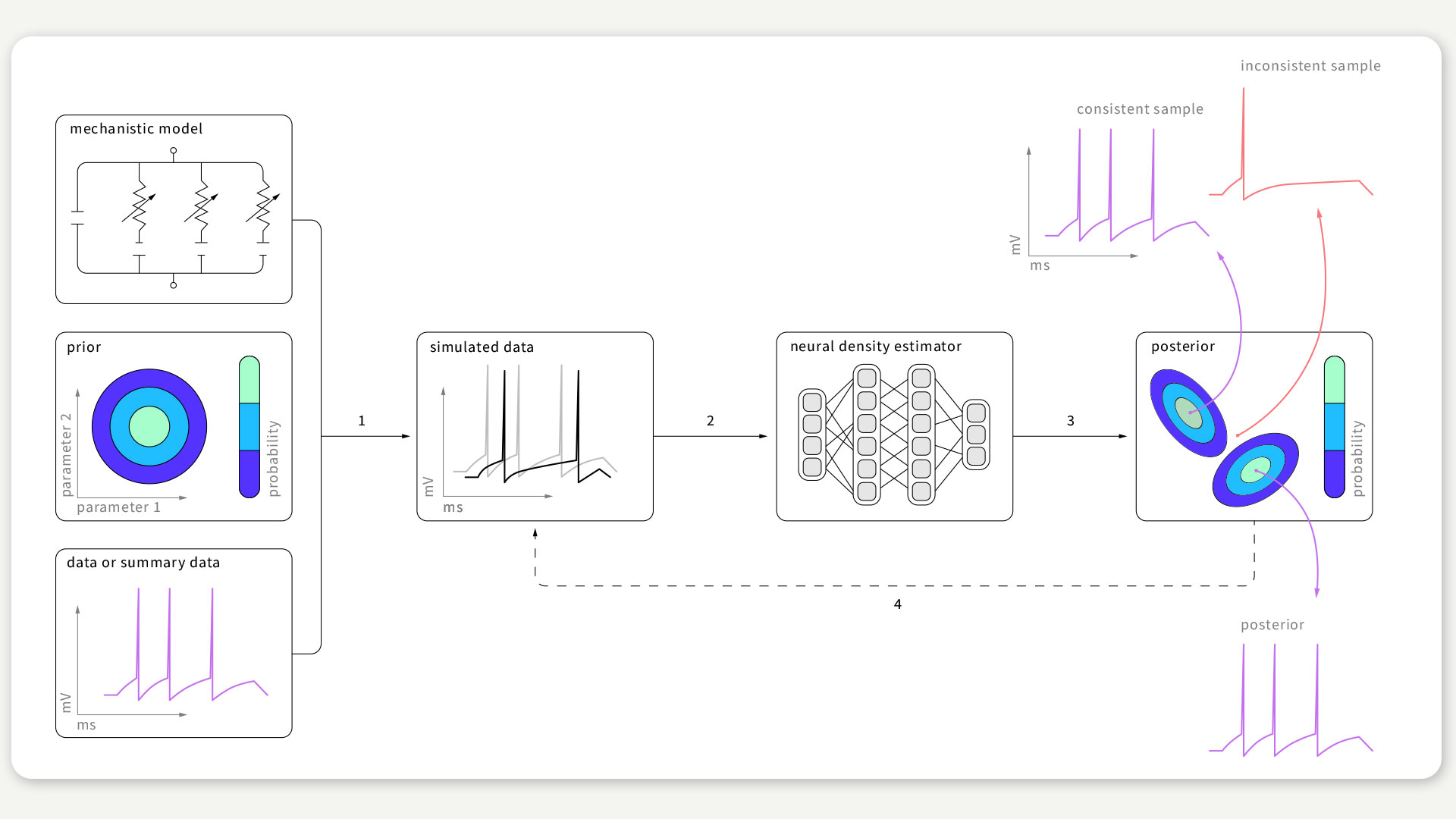
FIGURE 1 / The SNPE algorithm aims to find data-compatible models. It takes three inputs: a model (i.e. computer code to simulate data from parameters), prior knowledge or constraints on parameters, and empirical data. SNPE runs simulations for a range of parameter values, and trains an artificial neural network to map any simulation result onto a range of possible parameters. After training on simulated data with known model parameters, SNPE provides the full space of parameters consistent with the empirical data and prior knowledge. Figure according to Gonçalves et al., 2020.
From “trial and error” to an algorithmic identification of suitable models
In a publication led by Pedro Gonçalves, Jan-Matthis Lueckmann, and Michael Deistler from the lab of Jakob Macke, we tackled the problem of identifying suitable models. Up until now, the possibilities of finding suitable models were limited: Either models were modified in painstaking work, until they could explain the measured data, or attempts were made to heuristically match models by excessive trying – i.e. testing as many models as allowed by the compute resources. These approaches are either not systematic and prone to subjective criteria, or they are inefficient and hence limited by the complexity of the model.
To overcome these limitations, we developed an algorithm that automatically determines suitable models based on the data observed in the experiment. Key to this approach is “Bayesian inference”: Using the observed data and model simulations, the algorithm identifies all suitable models that fit the measured data.
Remarkably, this new approach does not require any knowledge of the internal workings of the model and hence can be applied to models of various kinds and complexities. We demonstrated the efficiency of the algorithm on several canonical models from neuroscience and suggest that the algorithm can be used in other areas of biomedical research as well.
Applying the new method: A model of retinal prosthetics to restore vision
In the lab of Philipp Berens, Jonathan Oesterle, Christian Behrens and others used this algorithm to infer model parameters of three neuron types of the retina. Besides the parameter inference itself, a major goal of this study was to bridge the gap between theoretical modelling and medical applications. For this, we created a model for the external electrical stimulation of the retinal neuron models, to simulate the retinal stimulation utilized in retinal neuroprostheses. We explored a variety of electrical stimuli and searched for stimuli selectively targeting either the ON or the OFF bipolar cell, which would help to better encode visual scenes into electrical signals and is currently one of the major problems of retinal implants.
For such a model-driven approach it is crucial to choose the model parameters carefully. We therefore based the initial parameter estimates of the modelled retinal neurons on information available in the literature. Since this data was scarce and not sufficient to constrain the model parameters, we used the SNPE-tool to further optimize the cell parameters, and obtained cell models matching previously recorded light responses recorded with two-photon imaging. Moreover, we used this algorithm to infer the parameters of the electrical stimulation.
Combining the cell models and the model for electrical stimulation, we were able to explore the effects of different stimulus settings in silico. After we validated our simulations with available data on stimulation thresholds for different stimulation settings, we tested a variety of electrical stimuli for selective ON and OFF bipolar cell stimulation. Our model suggests that triphasic currents might be able to selectively target the OFF bipolar cell due to its expressing of T-type Calcium channels. These channels open in response to relatively steep but brief increases in stimulation current, leading to calcium-activated neurotransmitter release. On the other hand, we did not find any stimuli that would exclusively stimulate ON bipolar cells.
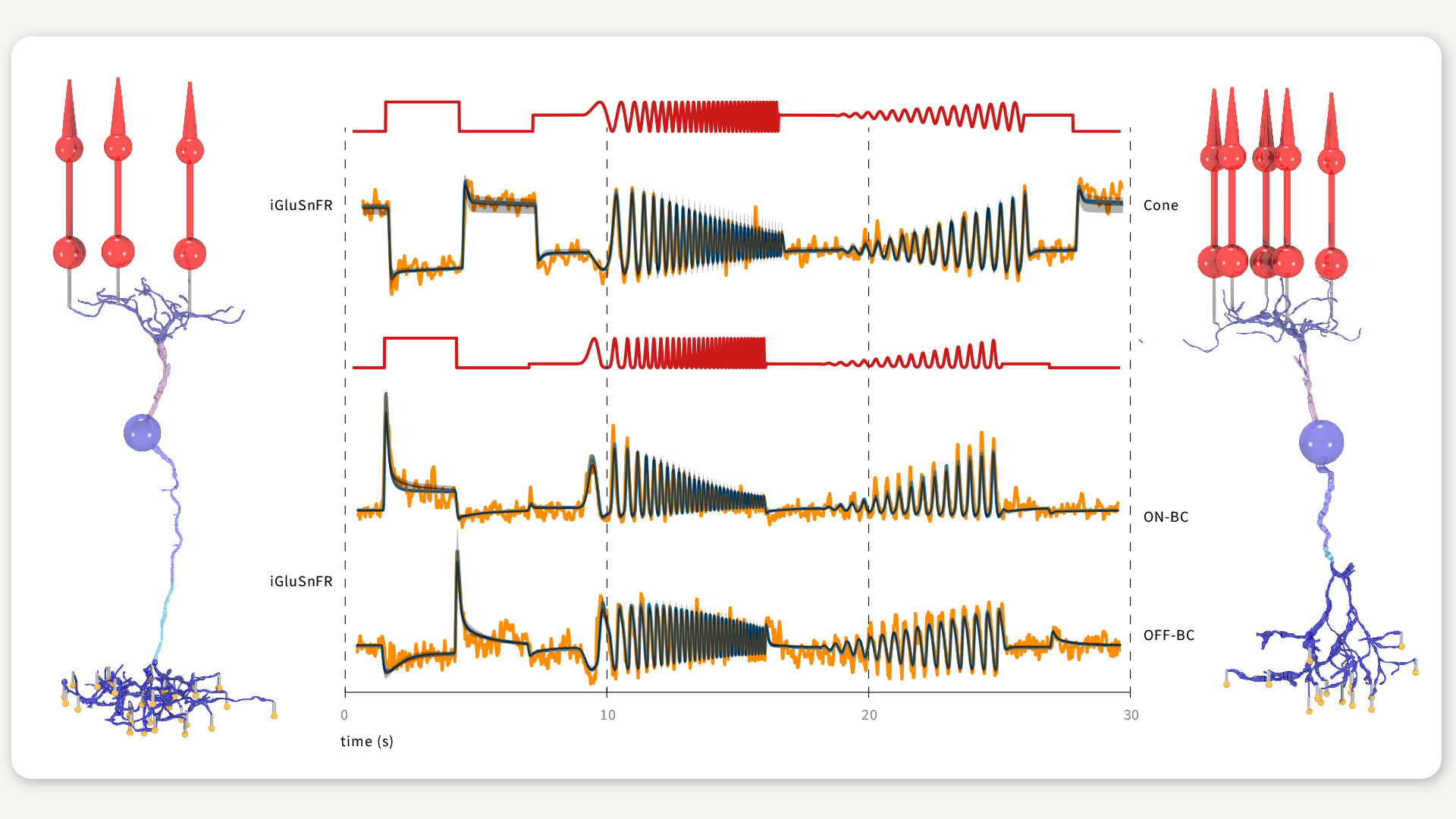
FIGURE 2 / PARAMETER INFERENCE FOR RETINAL CELL MODELS In the lab of Philipp Berens, we inferred the parameters of a cone and an ON (left cell) and OFF bipolar cell (right cell) receiving input from three and five cones (red cells), respectively. We fitted the model outputs (blue and black) to experimentally recorded responses (orange) in response to simple light stimuli (red traces) matching the experimental conditions. Figure according to Oesterle et al., 2020.
Conclusions
The two studies described above show how method development and application to challenging scientific problems can go hand-in-hand and complement each other. We believe that these findings are a first step in the quest to provide efficient neuroprosthetic devices for the blind, but also demonstrate the usefulness and challenges of the inference tool. In the future, this tool could be applied to a wide range of computational investigations in neuroscience and other fields of biology, which may help bridge the gap between “data-driven” and “theory-driven” approaches.
Original publications:
Pedro J Gonçalves, Jan-Matthis Lueckmann, Michael Deistler et al. Training deep neural density estimators to identify mechanistic models of neural dynamics. eLife 2020;9:e54997.
Text: Michael Deistler, Jonathan Oesterle
Cover illustration: Franz-Georg Stämmele

Using Machine Learning for 3D Soil Mapping
Comments