The brain has amazing capabilities: it recognizes complex visual patterns, controls movements, and can remember past experiences. But how does it do this? Advances in artificial intelligence (AI) in recent years have made it possible for artificial neural networks to perform similar tasks – thanks in part to powerful software that enables the efficient training of artificial neural networks. It was therefore hoped that artificial neural networks would provide insights into how the brain works. However, artificial networks are based on highly simplified neuron models, which is why they have so far made only a limited contribution to a better understanding of the brain. In contrast, there are also biophysically detailed brain models, but these are not capable of solving cognitive tasks such as recognizing visual patterns or storing memories.
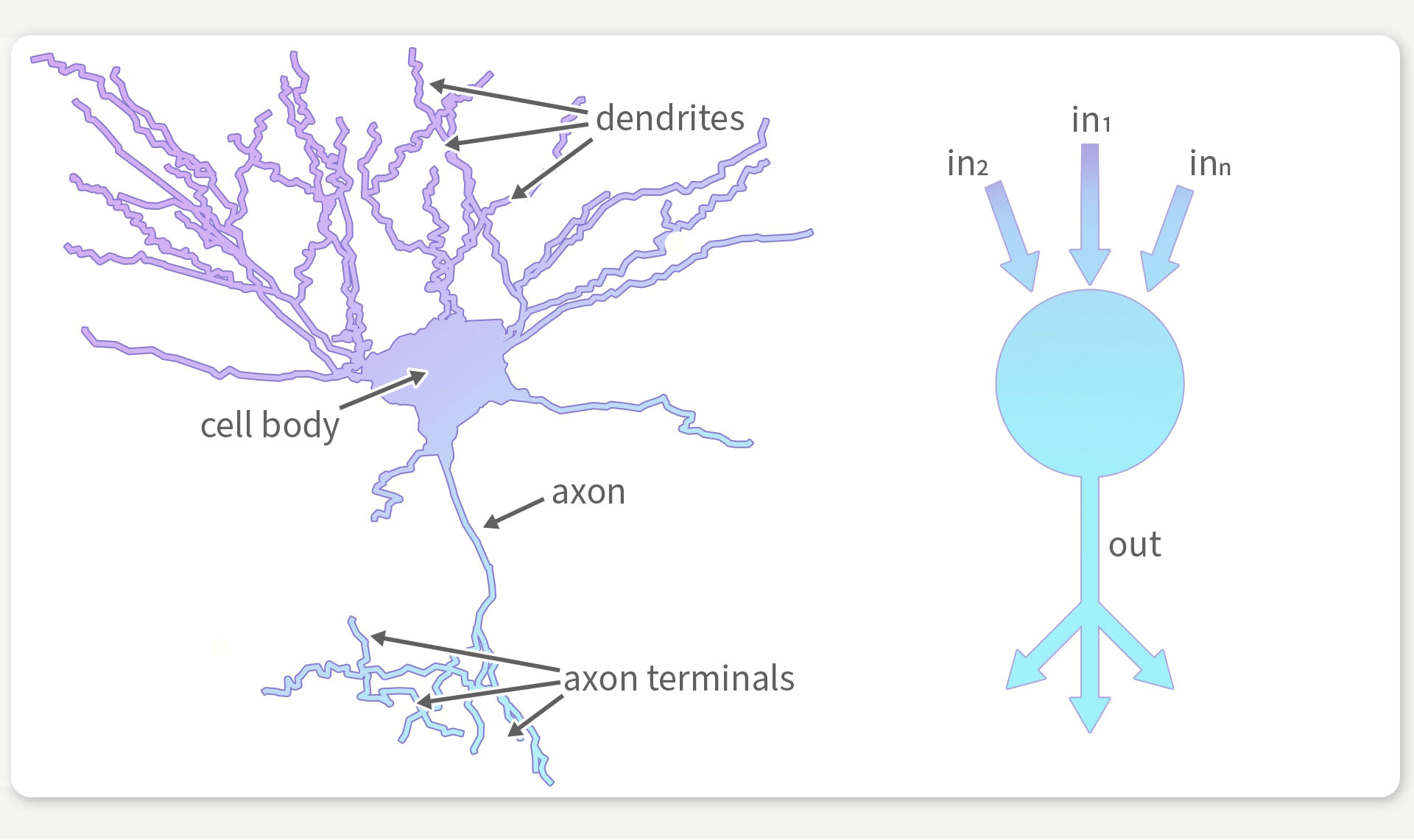
Nerve cells in the brain consist of a multitude of processes that influence the electrical activity of the cell (left). Modern AI architectures, on which previous brain simulations are often based, greatly simplify neurons (right), so that results often no longer allow conclusions to be drawn about the functionality of the brain. © Franz-Georg Stämmele
Training biophysically detailed brain models to solve cognitive tasks
Together with a team from the labs of Jakob Macke, Philipp Berens (University of Tübingen) and Pedro Gonçalves (VIB-Neuroelectronics Research Flanders, NERF), we have now developed a new software called JAXLEY. For the first time, this software makes it possible to train detailed models of the brain to solve cognitive tasks. JAXLEY thus lays the foundation for a new generation of brain simulations that will allow deeper insights into how the brain works and what it is capable of. The JAXLEY software and demonstrations of brain simulations that can be realized with it were recently published in Nature Methods.
Simulating brains with biophysically detailed neuron models
In 1963, Alan Hodgkin and Andrew Huxley won the Nobel Prize in Physiology and Medicine for describing how cellular processes lead to the “action potential”: the fundamental signal emitted by neurons of (almost) every animal. In the decades that followed, many neuroscientists extended the “Hodgkin-Huxley” model. They incorporated the morphology of individual neurons, the contributions of dozens of ions, and even the diffusion of ions within cells. To enable neuroscientists to handle and simulate such increasingly complex simulations, many toolboxes for biophysical modelling in neuroscience emerged – most notably, the NEURON toolbox. The NEURON simulator has been maintained for almost 50 years now, and the codebase is so large that it feels as if there is no biophysical detail that cannot be implemented.
However, the NEURON simulator has a major disadvantage: Since it is written in the programming language “C”, it cannot benefit from the capabilities of modern programming languages. In particular, modern deep learning libraries allow training models with backpropagation of error, a method which measures how each parameter contributed to a mistake and adjusts parameters to reduce future errors. They can also speed up training with GPUs (Graphics Processing Units, processors that can handle many calculations in parallel) – with virtually no effort from users. To bring these capabilities to biophysical simulations of the brain, we developed a new biophysics simulator: JAXLEY (JAX & [Hodgkin-Hux]ley).
Optimizing brain simulations through backpropagation of error
JAXLEY makes it possible to simulate biophysical models – just like the NEURON simulator. On top of this, however, it also enables backpropagation of error and large-scale GPU parallelization, and thereby enables training of biophysical neuron models, as if they were artificial neural networks. JAXLEY can train the size of the neurons, the strength of the connections, and the number of ion channels, such that the simulation matches experimental recordings of neural activity or such that it solves a cognitive task. All of this is achieved because JAXLEY is fully written in JAX. JAX is a deep learning framework which excels at speeding up long programs (for example, minute-long brain simulations) and at scaling simulations or training to multiple GPUs or TPUs.
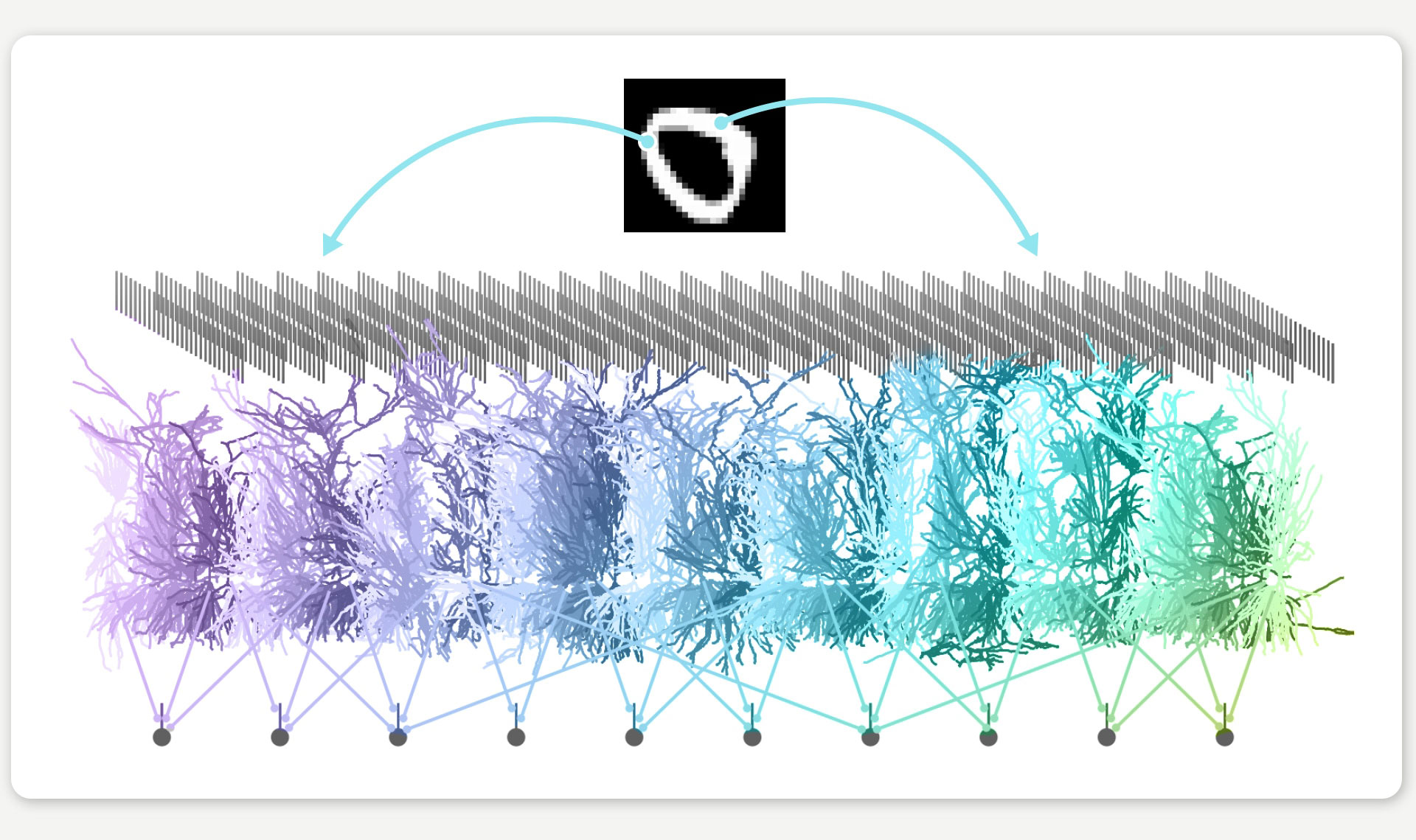
A brain simulation classifies a digit – in this case, a zero. Pixel values are fed into the network as input (illustrated here using two pixel values and the blue arrows) and processed step by step in several layers until the digit is correctly identified at the end. Each of these layers consists of hundreds of artificial neurons. JAXLEY makes it possible to simulate these artificial neurons realistically and solve the task at the same time. © Michael Deistler/Franz-Georg Stämmele
With the help of JAXLEY, our team built computer models of parts of the brain, starting with individual cells. We trained the biophysical processes within these virtual cells so that their activity matched that of real brain cells. In the next step, we built a model of a mouse’s retina and finally complex networks, which we trained with the new software to perform demanding tasks: The models were able to recognize visual patterns or store memories and retrieve them later.
Applying JAXLEY to investigate neural mechanisms and brain function
JAXLEY enables neuroscientists to investigate how neural mechanisms contribute to task solving. Although a single neuron is only micrometers in size, it contains tens of thousands of ion channels that influence the electrical activity of neurons. In the future, JAXLEY will allow researchers to explore the complexity of the brain and map it in computer simulations. In the long term, such simulations could also be used in medicine, for example to better understand neurological diseases or to investigate the effects of drugs virtually in advance.
Original publication: Michael Deistler, Kyra L. Kadhim, Matthijs Pals, Jonas Beck, Ziwei Huang, Manuel Gloeckler, Janne K. Lappalainen, Cornelius Schröder, Philipp Berens, Pedro J. Gonçalves, Jakob H. Macke: Jaxley: Differentiable simulation enables large-scale training of detailed biophysical models of neural dynamics, Nature Methods, 2025. https://doi.org/10.1038/s41592-025-02895-w
Cover illustration: Franz-Georg Stämmele

Scholar Inbox: Daily Research Recommendations just for You

Comments